Welcome!

Welcome to my website. I am a Senior Population Geneticist, with a Ph.D. in evolutionary biology, and broad multi-species interests in herpetology, population genetics, genomics, evolutionary ecology, and human evolution. Specifically, I am interested in the process of environmental adaptation, how it proceeds at the genomic level, and how it applies to species and population persistence. My research has unraveled ancient population history in Yosemite toads (Anaxyrus canorus), demonstrated adaptive differences in their larval development rates, and suggested that secondary contact zones could catalyze genetic adaptation to climate change. I have also developed numerous tools for human genetic genealogical research: myOrigins® 3.0, the Chromosome Painter, Big Y Age Estimates, the FTDNATiP™ Report, the upcoming Mitotree, Geo-Genetic Triangulation (for Beethoven research), and now Globetrekker.
Check back soon to see more about my research, fieldwork, latest publications, programming, and wildlife/nature photography!
January 2025
The Amphibian Population Task Force (2025) meeting was a huge success! Over 200 participants, which is a new record. Jeff Mabe and I gave the “big reveal” about hybrids between two species, Western and Yosemite Toads. We also had a great time catching up with old friends, and planning our upcoming research!

November 2024
The hybrid panel is finished! After almost 70 years, we now know whether or not Yosemite and Western Toads hybridize near Lake Tahoe, check it out!
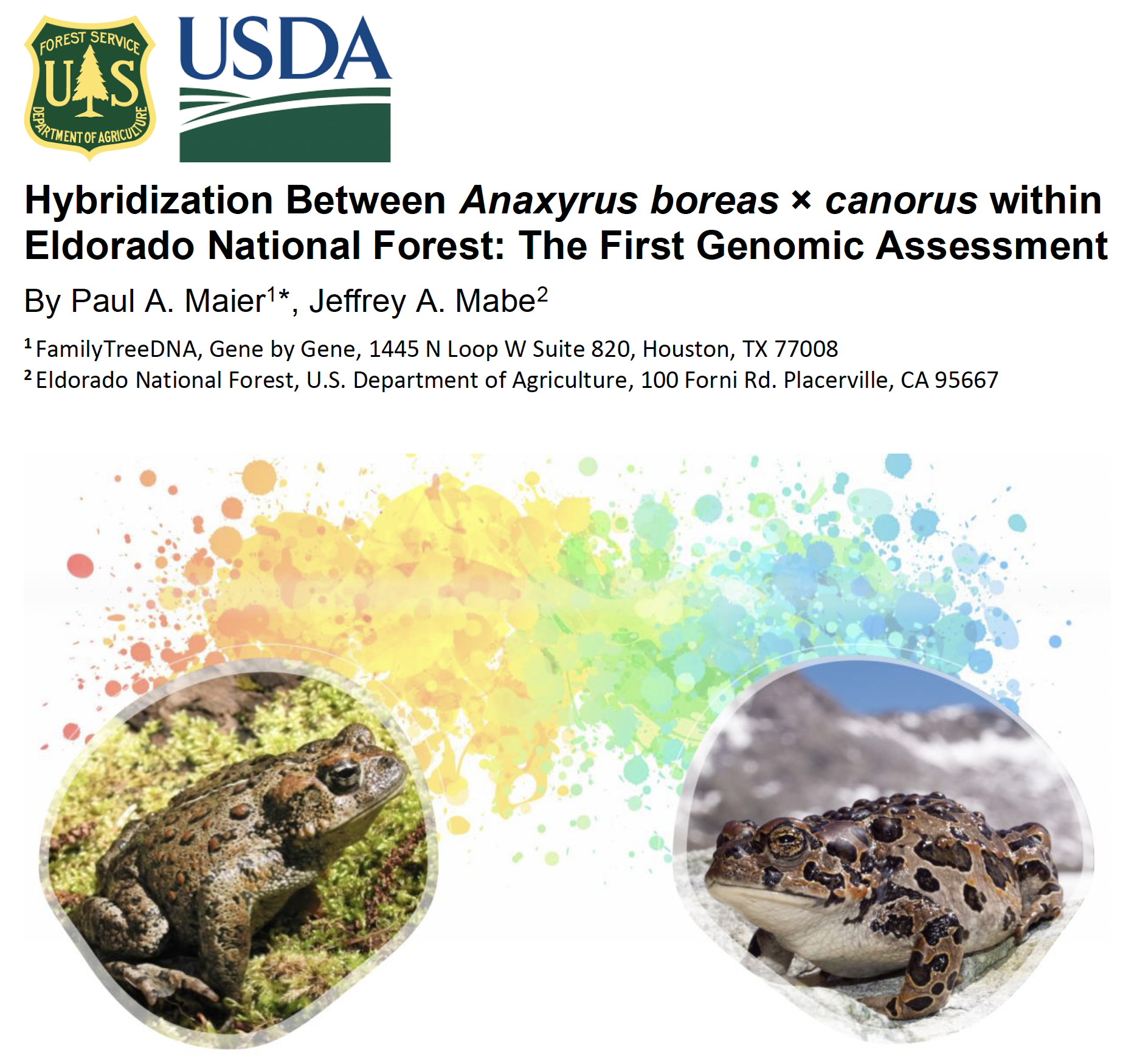
Maier P.A., Mabe J.A. (2024). Hybridization between Anaxyrus boreas × canorus within Eldorado National Forest: The first genomic assessment. Final Report to US Forest Service. Eldorado National Forest, Placerville, CA. [PDF]
March 2024
Tucson Herpetological Society invited me to give a talk about Yosemite toads, which my wife and daughter attended, along with a few old friends!
March 2024
Big news! After 5+ years and 5 publications, my entire Ph.D. is published!
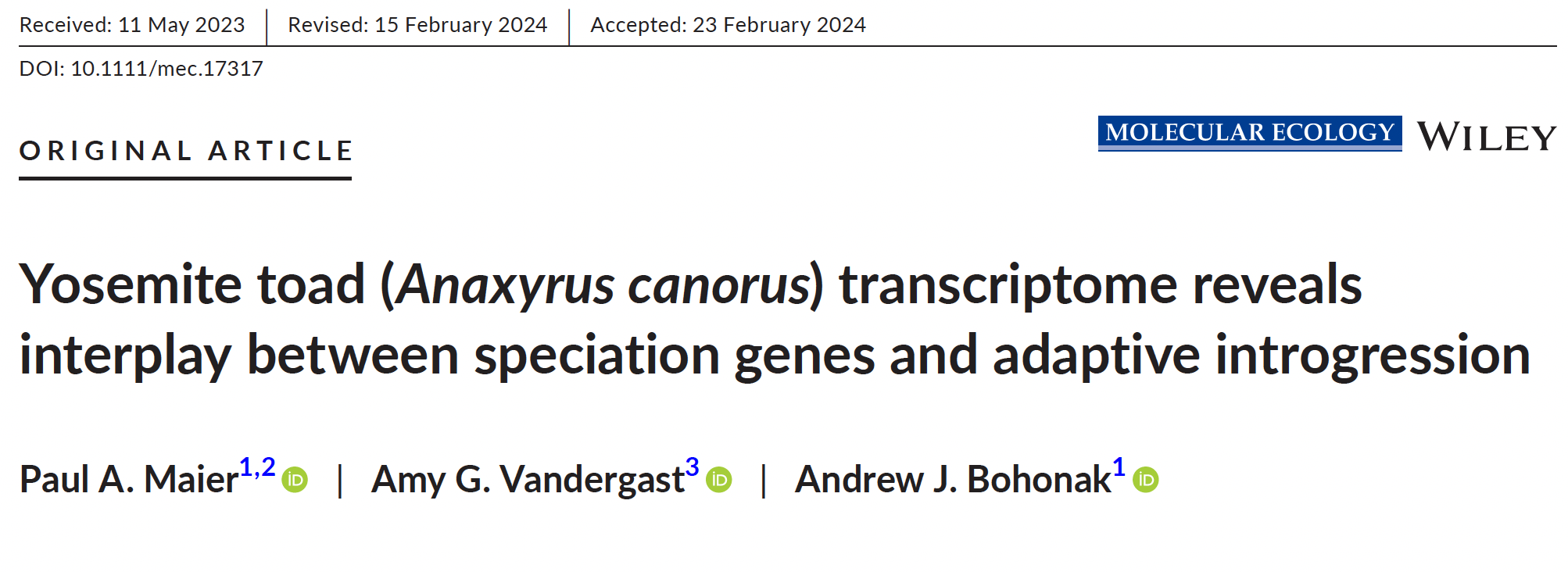
This paper was a major achievement, involving the assembly of an entire transcriptome (all protein-coding sequences) for a species lacking any genomic resources.
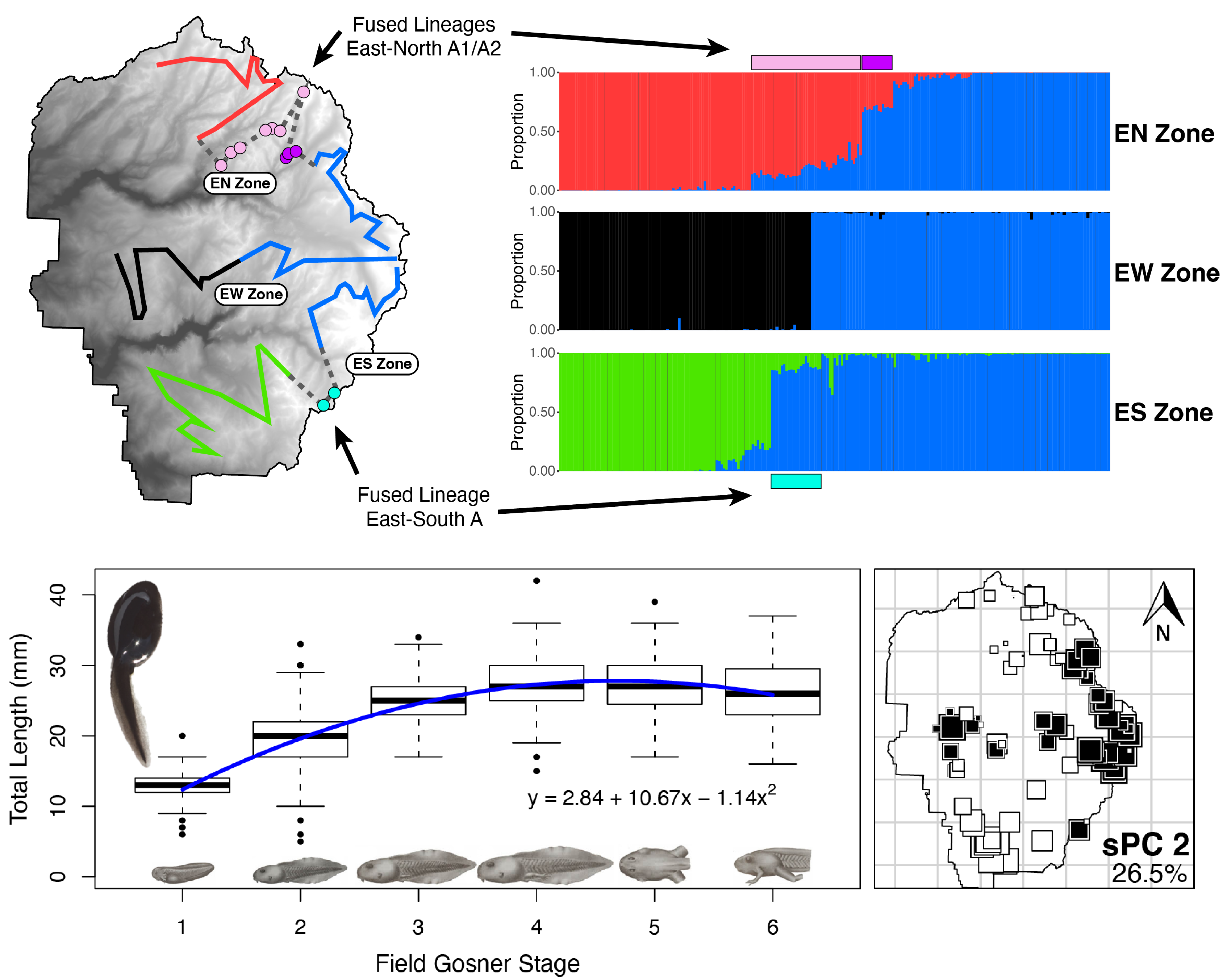
We used 3 Yosemite toad hybrid zones as 3 independent replicates to ask the question: How predictable is adaptation in nature? We looked at two different types of adaptations: (1) “adaptive divergence” genes helping lineages to separate, and (2) “adaptive introgression” genes helping lineages to reconnect and spread beneficial genes with each other. These two ideas may at first seem conflicting, but we found they are much more compatible than currently appreciated. We also found predictability, not just in terms of which genes are repeatedly used, but their functional pathways (different genes but similar purposes). Our newly built transcriptome revealed that one of these genetic loci is a lipid metabolism gene potentially involved in climate change adaptation.
Maier P.A., Vandergast A.G., Bohonak A.J. (2024). Yosemite toad (Anaxyrus canorus) transcriptome reveals interplay between speciation genes and adaptive introgression. Molecular Ecology. 33(8), e17317. [DOI] [PDF]
February 2024
Just back from RootsTech 2024 in SLC, Utah—FamilyTreeDNA had a fantastic showing! Several families I talked to had major epiphanies as we talked through their Discover™ results. It feels amazing to help people break down their brick walls! After talking to hundreds of customers, two things were clear: (1) Everyone is hyped about Globetrekker, and (2) Everyone is brimming with anticipation for the upcoming release of Mitotree!
Here are several photos from our booth…



November 2023
I’m back after a long hiatus… The most important event in my life took place over the summer: My daughter was born 🥰😍🥰😍🥰
Immediately after returning, FamilyTreeDNA had its first conference since before the pandemic (2019)! I gave this virtual presentation for the group administrators, explaining the beauty of our new Y-DNA Time Tree, and our amazing phylogeographic tool Globetrekker.

September 2023
Here’s the 3rd and final installment of our blog series about Globetrekker! Miguel Vilar dives into the anthropology underpinning these newly charted human migration paths.
Globetrekker, Part 3: We Are Making History
August 2023
Here’s the 2nd installment of our blog series about Globetrekker—written by yours truly! This is “How it’s Made” and talks about ice sheets, sea levels, and the phylogeographic science behind this amazing new tool!
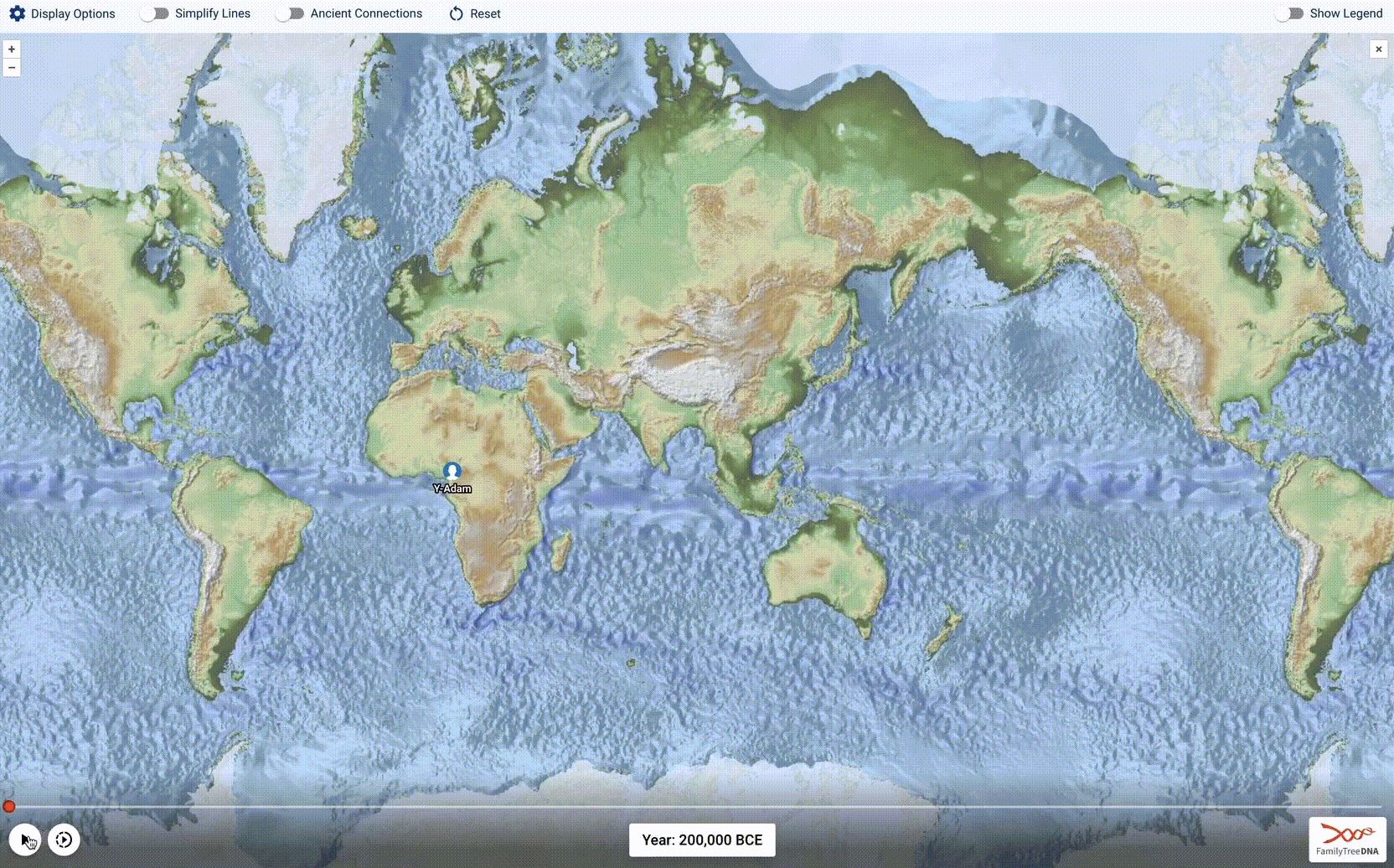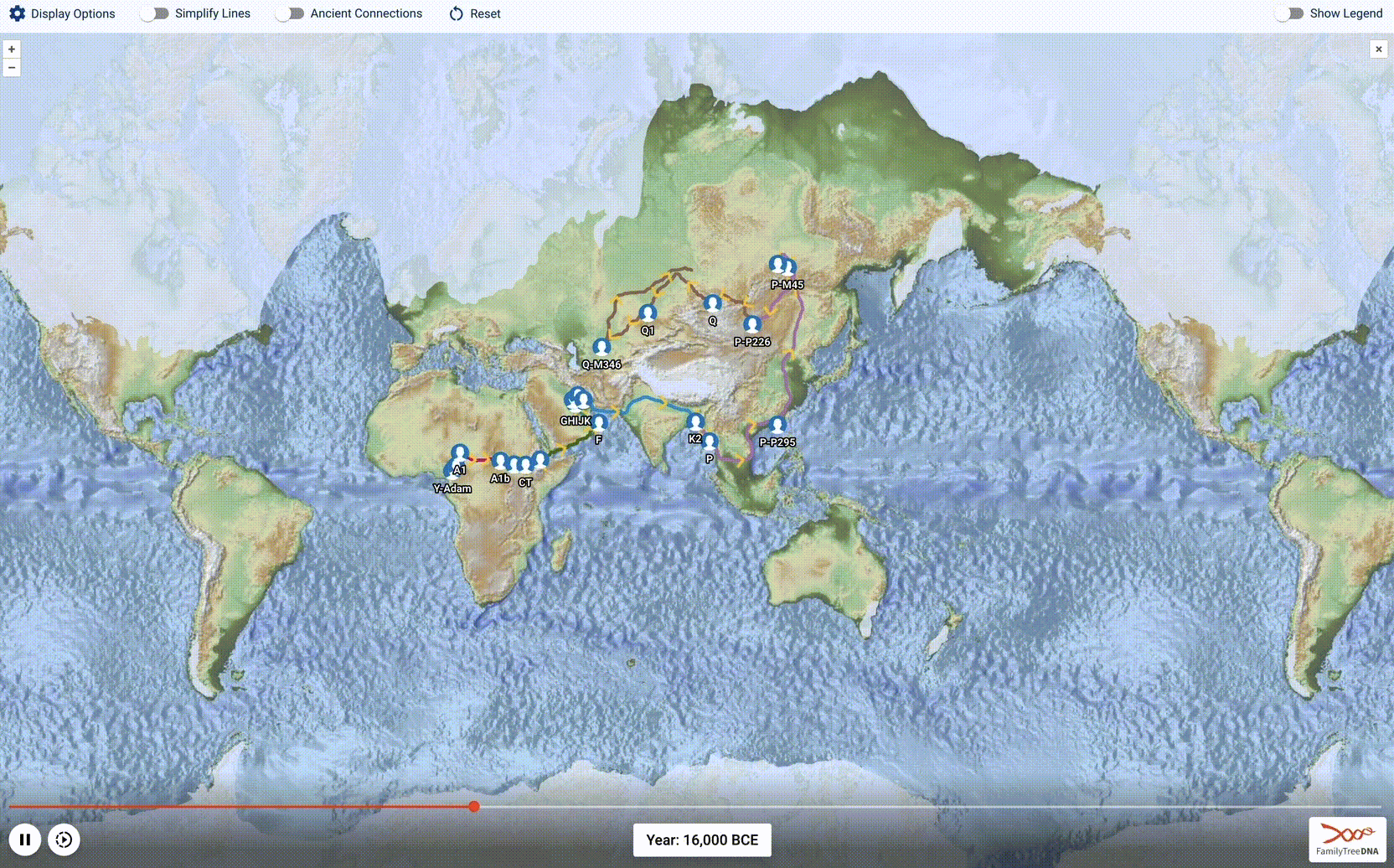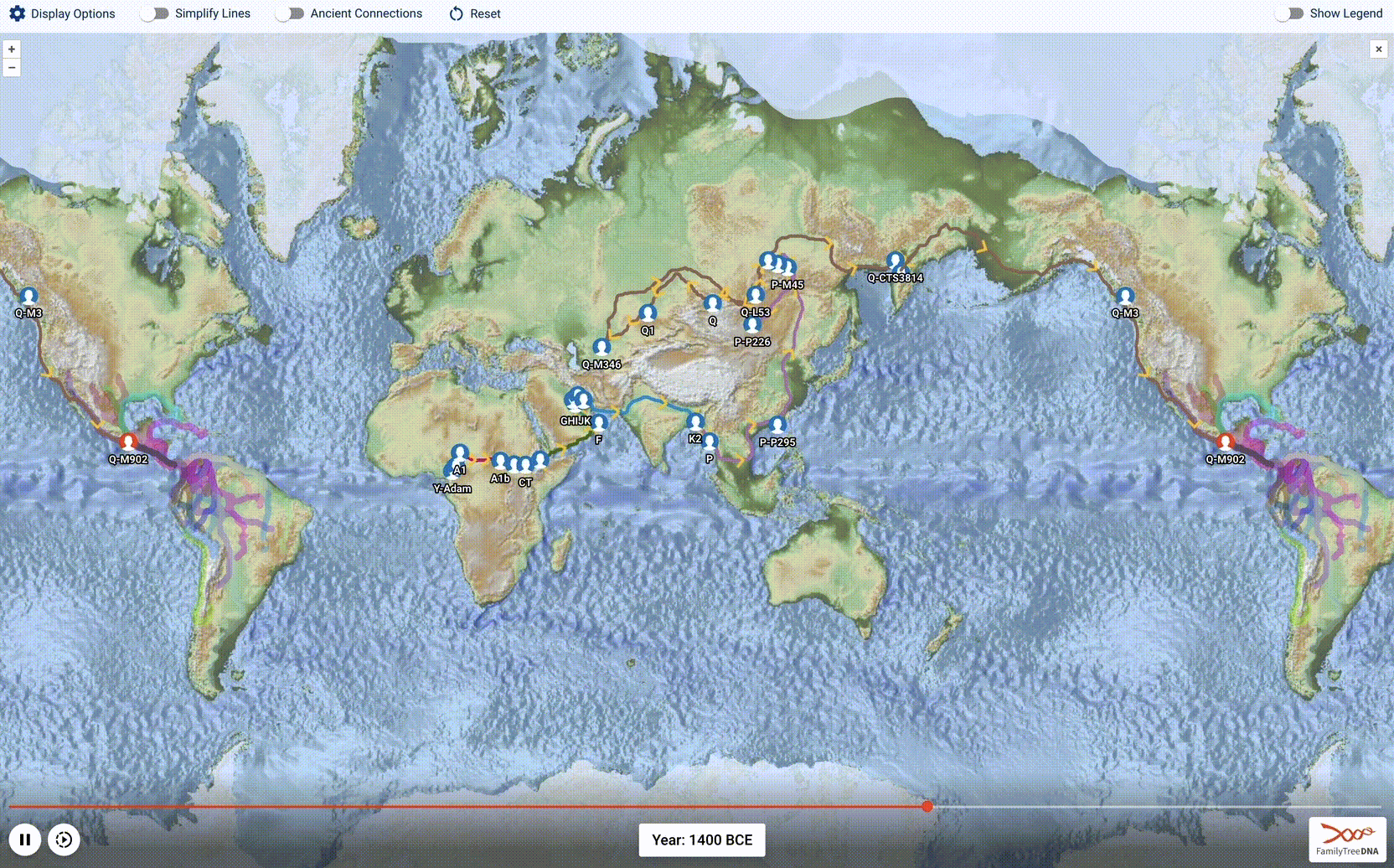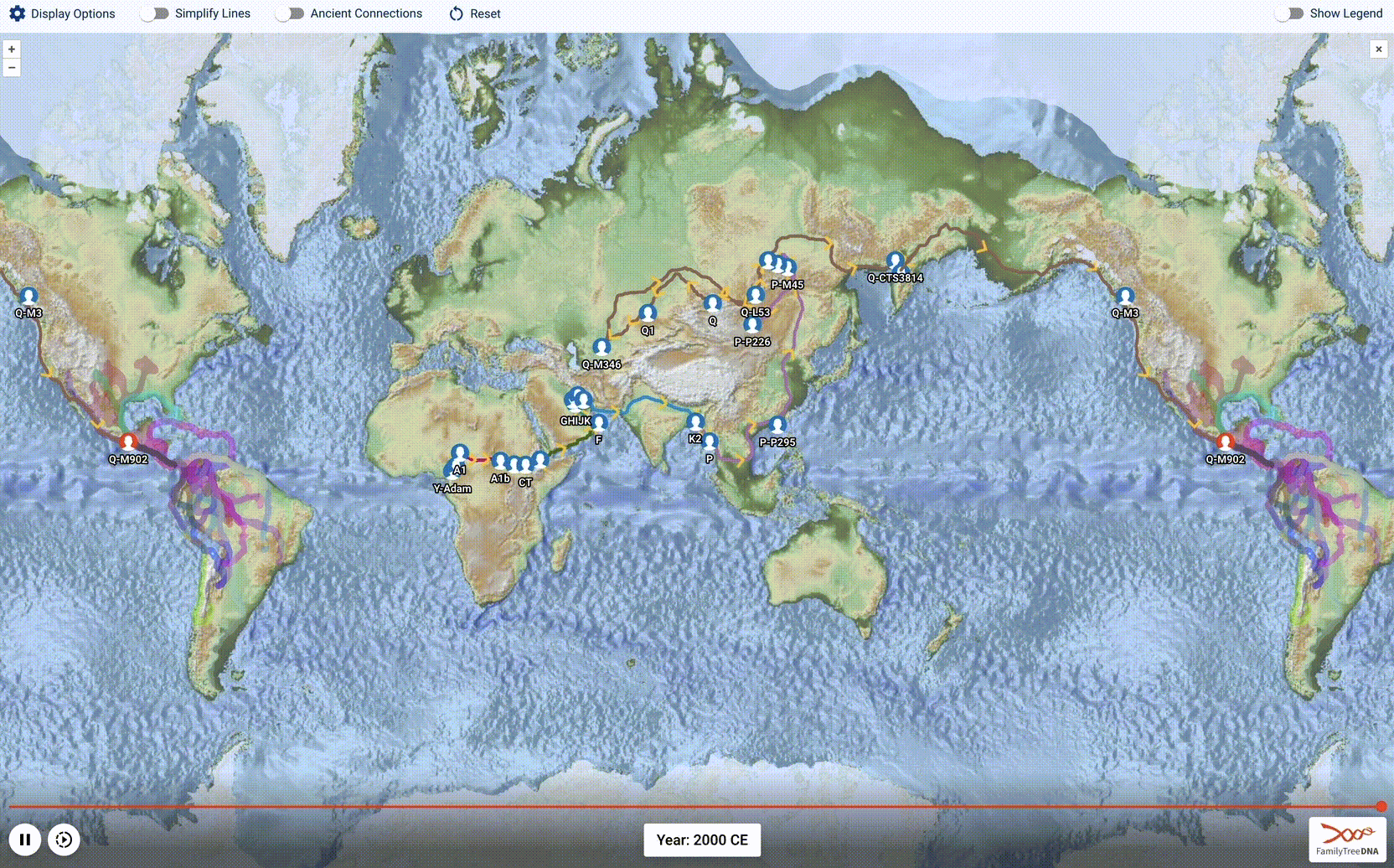
Globetrekker, Part 2: Advancing the Science of Phylogeography
July 2023
After a year of work, we launched an amazing new tool: Globetrekker! This is the 1st installment of our blog series describing how we charted >50,000 human migrational paths across the planet, starting in the cradle of humanity 200,000 years ago and ending in YOU.
Globetrekker, Part 1: A New FamilyTreeDNA Discover™ Report That Puts Big Y on the Map
June 2023
Had the pleasure of sitting down with Chelsea Rose of NPR Jefferson Public Radio’s Jefferson Exchange! We had a great chat about the Beethoven study, and the revelations about history that we can squeeze out of just a lock of hair.

May 2023
The ARCS Foundation, who partially funded my Ph.D. work, did a nice interview on my recent research at FamilyTreeDNA: including the 100,000-year-old L7 discovery, Beethoven’s genome, and myOrigins!

April 2023
Here’s a little presentation I gave at FamilyTreeDNA’s town hall about the genomics of Ludwig van Beethoven—our analysis of his DNA has permanently altered our understanding of this quintessential composer.
March 2023
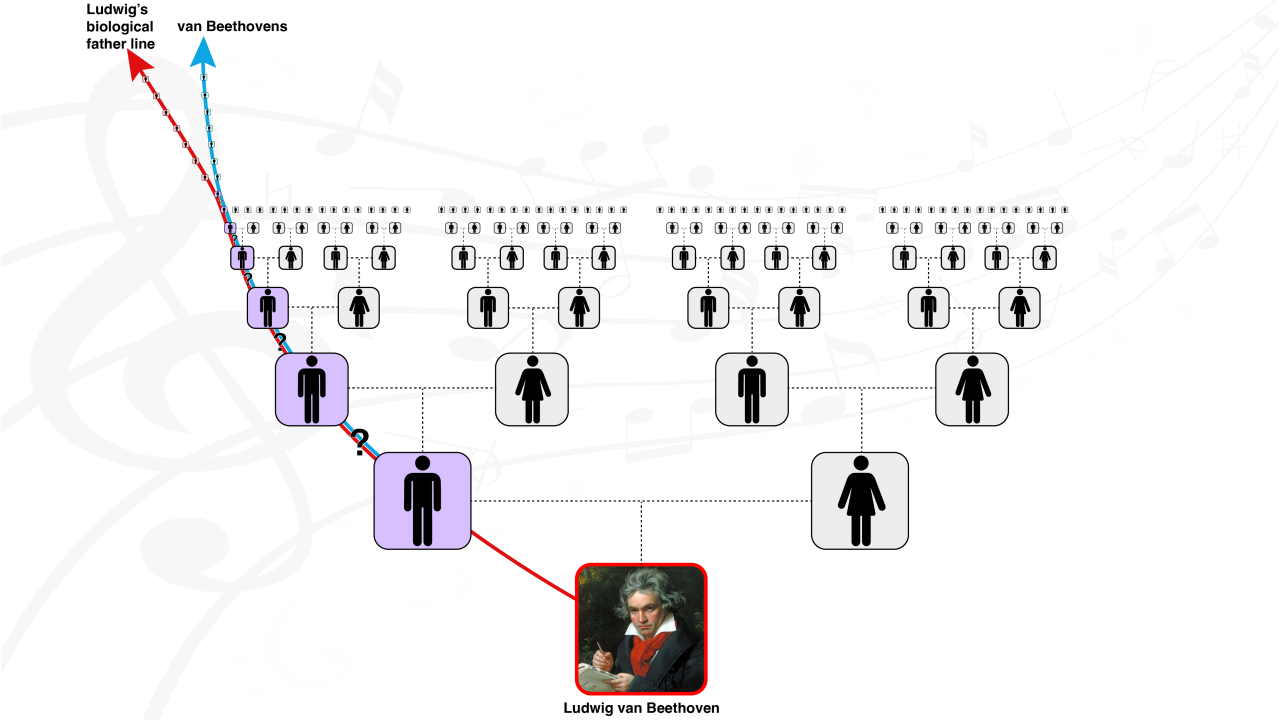
Here is a nice synopsis of our research on Beethoven!
March 2023
Big news! After three years of secrecy, we just published our genetic exposé of Ludwig van Beethoven!
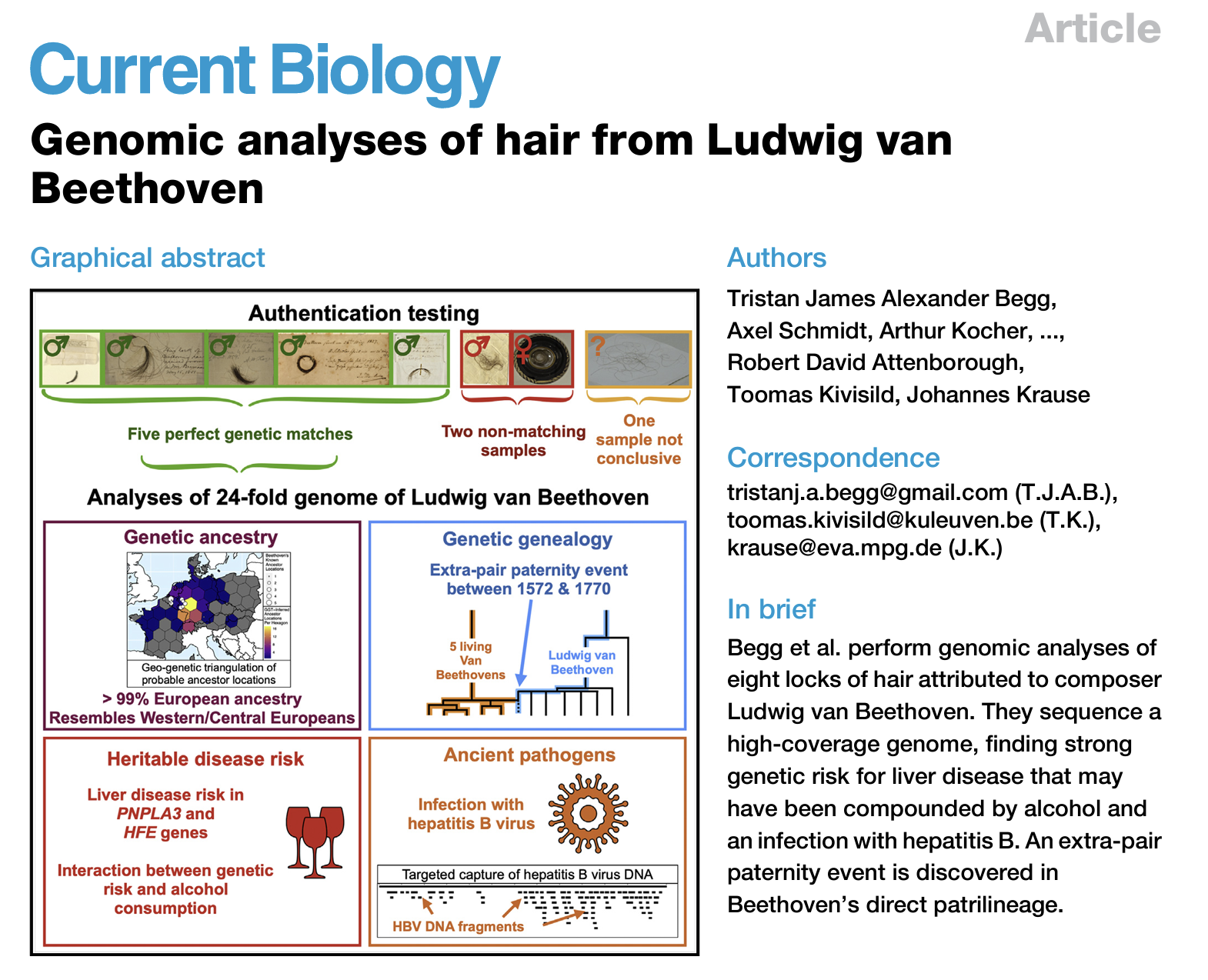
This follows a collaboration between FamilyTreeDNA, Cambridge University, the Max Planck Institute, and others to sequence Beethoven’s complete genome. The highlights of the study include: (1) the most famous Beethoven (Ludwig) was not genetically a Beethoven, (2) a genetic predisposition to liver disease coupled with hepatitis B possibly may have precipitated his early death, and (3) he was in a high percentile risk category to develop lupus, which can be associated with deafness.
My major contribution was to develop a novel method (Geo-Genetic Triangulation; GGT) to help validate that the sample indeed belonged to Beethoven.
Begg T.J.A., Schmidt A., Kocher A., Larmuseau M.H.D., Runfeldt G., Maier P.A., Wilson J.D., Barquera R., Maj C., Szolek A., Sager M., Clayton S., Peltzer A., Hui R., Ronge J., Reiter E., Freund C., Burri M., Aron F., Tiliakou A., Osborn J., Behar D.M., Boecker M., Brandt G., Cleynen I., Strassburg C., Prüfer K., Kühnert D., Meredith W.R., Noethen M., Attenborough R.D., Kivisild T., Krause J. (2023). Genomic analyses of hair from Ludwig van Beethoven. Current Biology, 33, 1–17. [DOI] [PDF] [PR1] [PR2] [NYT] [Video]
February 2023
Gene by Gene’s laboratory very kindly invited me to speak about population genetics—here’s a little presentation I call “The Sampler Platter,” about the history of the science, and all the cool facets of human evolution it has revealed.
January 2023
The Amphibian Population Task Force (APTF) meeting in Sebastopol, California, was a fantastic conference! I had the pleasure of seeing many old friends and colleagues spanning my amphibian career with the US Forest Service, US Geological Service, and UC Santa Barbara. I summarized over a decade of my research on Yosemite toads, which was a fun challenge.
Click these links to see the agenda and abstracts.
This talk followed the temporal framework of my dissertation, and recent publications:
- Ancient Past (phylogeography)
- Recent Past (adaptive introgression)
- Present Day (population structure)
- Near Future (range shifts, future adaptive conservation units)
December 2022
We just published the first ever landscape genomics study of Yosemite toads! What is landscape genomics? Essentially, it’s the science of discovering how species adapt to their environment, via the lens of genomic loci under natural selection, and using clever spatial methods.
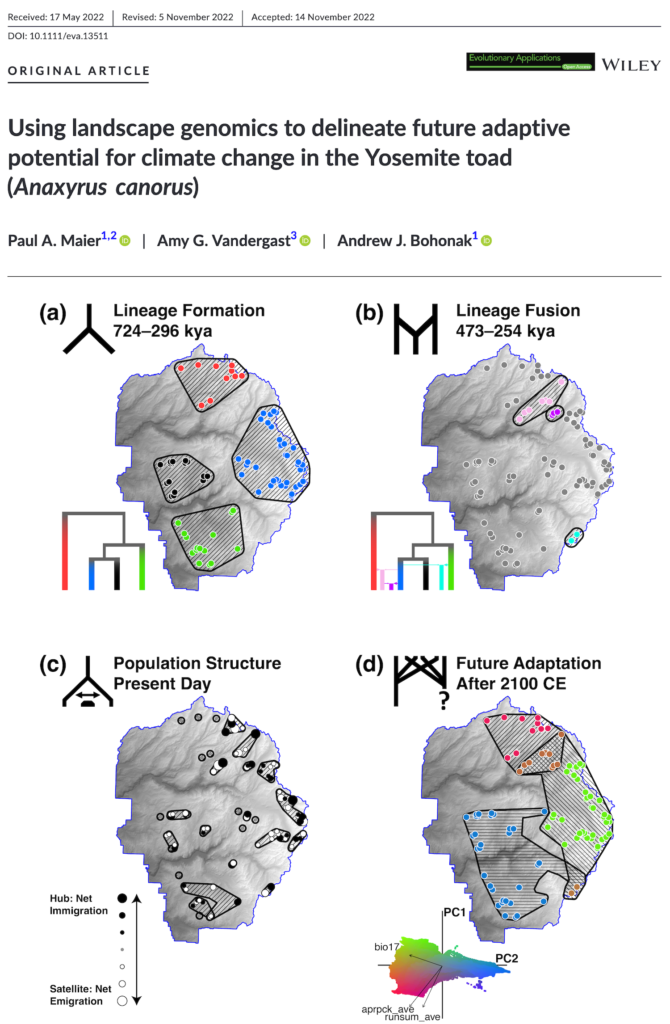
Not only that, but we made the case for looking at conservation using an explicitly future-directed approach. (Most conservation units are based upon past or present patterns.) In the future, we predict three lineages will emerge in Yosemite National Park based upon adaptation to three different climatic regimes (high snowpack, high summer rainfall, low precipitation). Using machine learning and geo-genomic simulations, we forecast a 29% species-wide decline over 90 years, however the “YF-East” lineage is most likely to persist into the future. That’s because it has the highest supply of climate-associated genomic diversity, and will have the smallest demand of climate-associated selection, in terms of required allele frequency shift!
Maier P.A., Vandergast A.G., Bohonak A.J. (2023). Using landscape genomics to delineate future adaptive potential for climate change in the Yosemite toad (Anaxyrus canorus). Evolutionary Applications, 16(1), 74–97. [DOI] [PDF]
October 2022
Our article made the cover of Heredity!
WE MADE THE COVER OF HEREDITY!!! 🧬🥳🧬🥳🧬 https://t.co/BtCYCFZsi0
— Dr. Paul Maier (@_Mountain_Man) October 30, 2022
The Heredity podcast with James Burgon did a great interview that summarizes our article. Well worth a listen!
August 2022
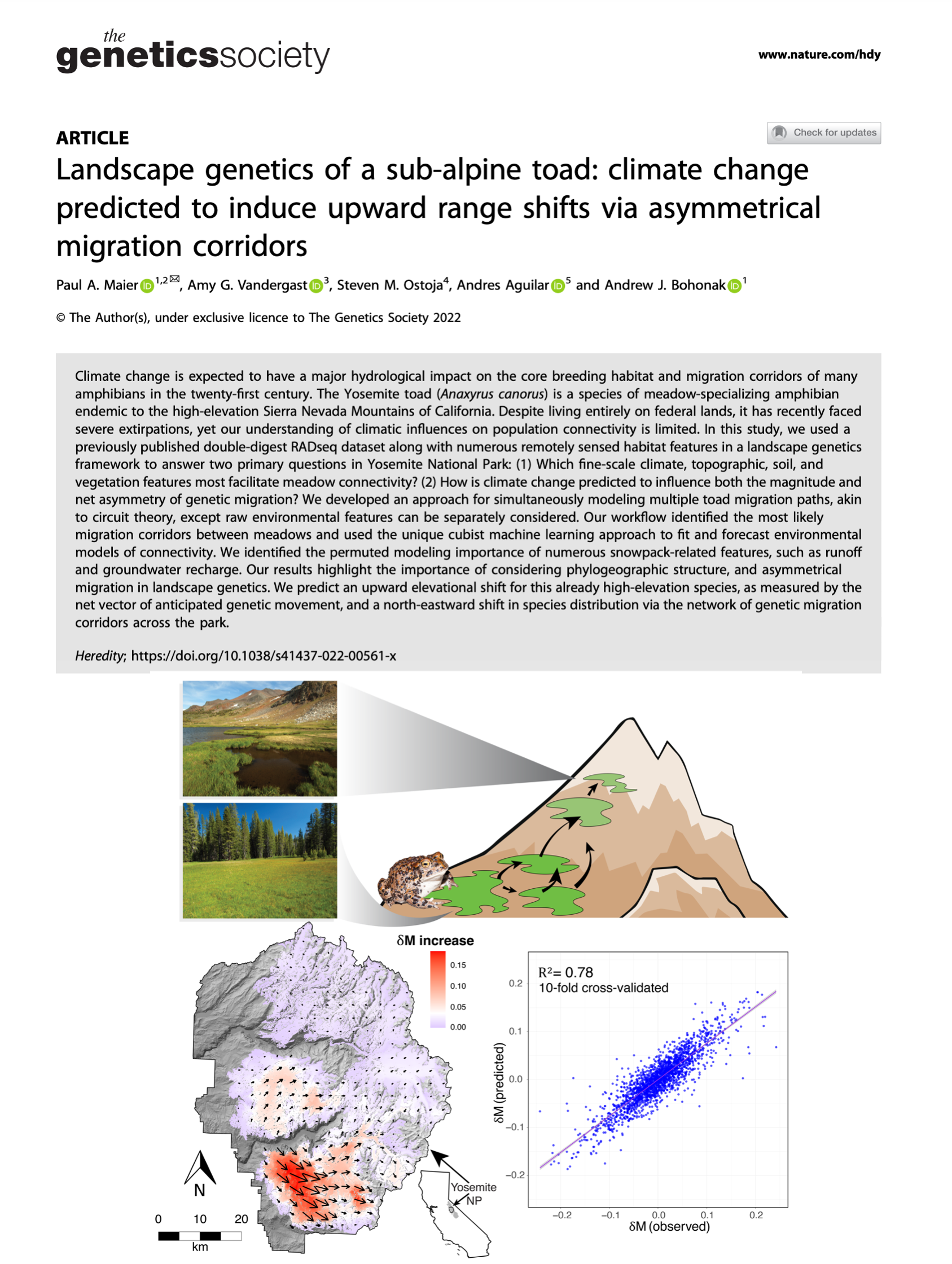
The rest of my third dissertation chapter is now published! We combined network methods and unique machine learning techniques to forecast that toads are shifting their range upward, due to climate change. (They are already sub-alpine!) Published in Heredity, a journal started by Ronald Fisher, who essentially invented my career of Population Genetics!
Maier P.A., Vandergast A.G., Ostoja S.M., Aguilar A., Bohonak A.J. (2022). Landscape genetics of a sub-alpine toad: Climate change predicted to induce upward range shifts via asymmetrical migration corridors. Heredity, 129(5), 257–272. [DOI] [PDF] [SharedIt] [Podcast] [Cover]
June 2022

The Million Mito Project team has just published their first major article! 100,000 years ago, when everyone’s ancestors (yours and mine) were African, severe volcanic eruptions and drought forced a few East African people to migrate into the African Great Lakes region. Their maternal lineage is the oldest found in 20 years! Yet it remained unknown for many reasons. One of those reasons is its rarity: only 19 out of hundreds of thousands of people examined inherited this legacy, which we call “L7”. In fact, one L7 participant in the study is 80,000 years removed from their closest relative, a new world record! Today, the Sandawe foragers of Tanzania boast the largest L7 percentage, but even they are only ~1% L7. Frankly, it’s amazing we could piece together this story, given that it takes place at the dawn of humanity. Imagine a time when our ancestors were inventing early fishing technologies, painting with the first ochre pigments, and our tongues were uttering the first click-based words. Mitochondrial DNA was at the heart of this discovery, and I frankly can’t wait to see what other stories it can reveal!
Maier P.A., Runfeldt G., Estes R.J., Vilar M.G. (2022). African mitochondrial haplogroup L7: A 100,000-year-old maternal human lineage discovered through reassessment and new sequencing. Scientific Reports, 12(1), 10747. [DOI] [PDF] [Video]
We talk about the paper more in this video:
We have a FamilyTreeDNA blog post about it:
A 100,000-Year-Old Human Lineage Rediscovered
And also a DNAeXplained blog post:
Mitochondrial Eve Gets a Great-Granddaughter: African Mitochondrial Haplogroup L7 Discovered
April 2022
We had a great time presenting at the East Cost Genetic Genealogy (EECG) 2022 conference, based (virtually) in Baltimore MD. Our 3 1/2 hour tag team presentation “mtDNA Academy” had surprisingly high attendance, with half of the participants joining … and (yet more surprising) … staying! We had a lot of fun breaking down the science, and the genealogy involved. My second talk about myOrigins 3.0 (MO3) showed some use cases for population segments, for example how small Native American segments can help confirm a suspected link between two cousins.


March 2022
Part of my third dissertation chapter is now published! We found that meadows are the most distinct unit of population (within the hierarchy of levels: meadow < meadow neighborhood < lineage < park). However, the story doesn’t end there. Neighborhoods of adjacent meadows are clustered across the landscape, and in many cases, consist of large flat “hubs” surrounded by smaller, and topographically complex, “satellites.” We also found that having adjacent upland (foraging/hibernating) habitat, and being closer to ridgelines, is associated with stronger hubs.
Maier P.A., Vandergast A.G., Ostoja S.M., Aguilar A., Bohonak A.J. (2022). Gene pool boundaries for the Yosemite toad (Anaxyrus canorus) reveal asymmetrical migration within meadow neighborhoods. Frontiers in Conservation Science, 3, 851676. [DOI] [PDF]
March 2022
RootsTech is here again!
Check out my “Innovations” talk about Age Estimates for our Big Y (paternal) haplotree:
And check out my classroom session about the Million Mito Project here:
February 2022
The Million Mito Project is live! Check out FamilyTreeDNA’s page here for more info:
https://www.familytreedna.com/mtdna-million-mito-project

September 2021
I’m very excited to release some FamilyTreeDNA videos, to accompany the recent release of the myOrigins 3.0 white paper, and the Chromosome Painter.
Part 1: An introduction to the feature:
Part 2: A discussion of the methods:
Part 3: Why don’t my results contain ____? A deep-dive into why population boundaries in Europe (and other places) are fuzzy.
August 2021
Our Research & Development Team at FamilyTreeDNA just released two monumental white papers!
The first is about the new myOrigins 3.0, which I am particularly proud of. This is more than a simple product description. It plunges the depths of what populations are, how they diverge in genomic space, and what tradeoffs exist when trying to estimate ancestry from them.
Here is one excerpt, representing the topology of major human lineages:
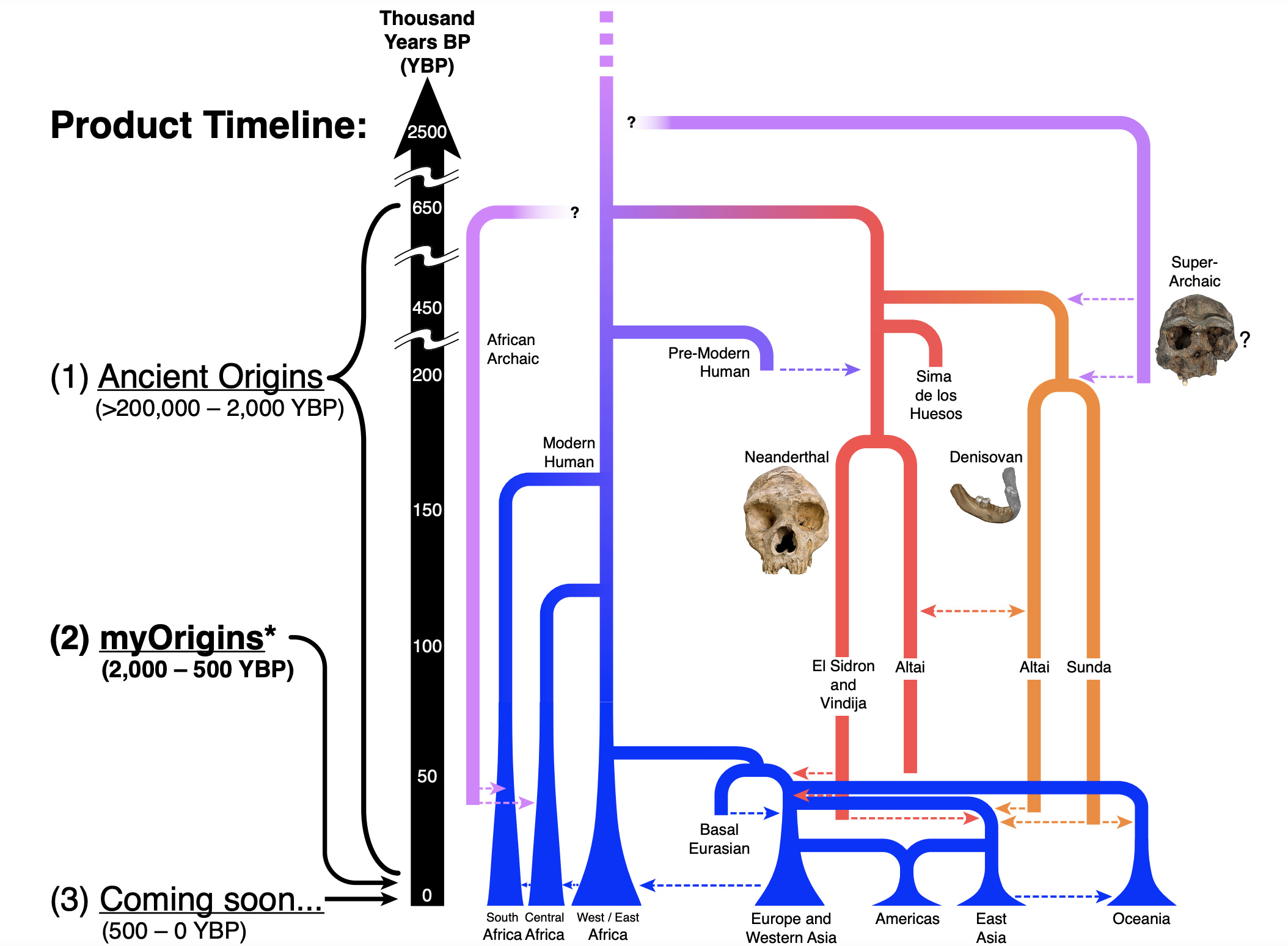
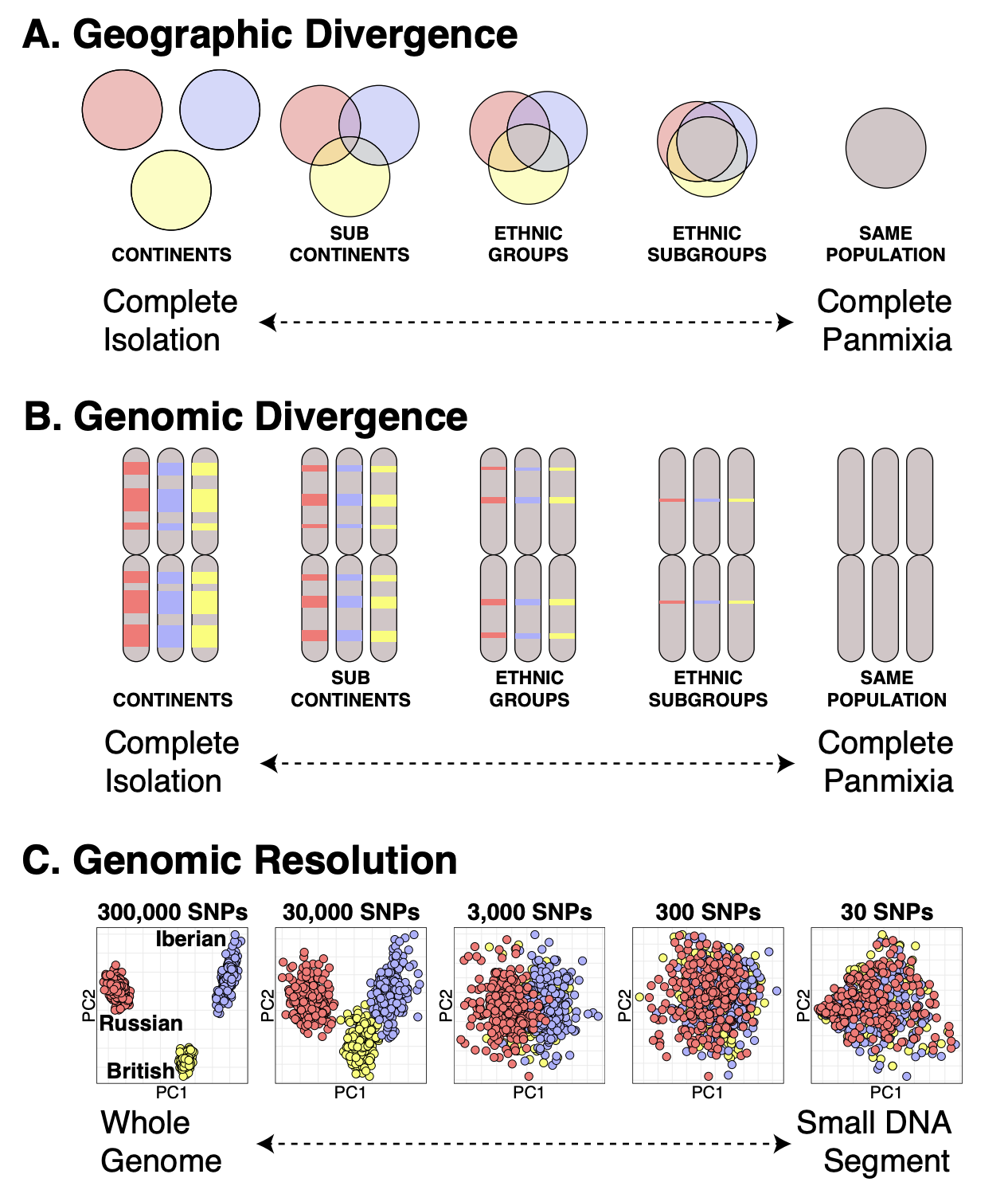
Here we can see a cartoon representing the spectrum of population distinctiveness: from complete isolation (requiring more time apart) to complete panmixia (individuals are equally likely to interbreed). Notice how genomic separation tracks geographic separation, with small but growing “islands” of divergence forming over time, which contain Ancestrally-Informative Markers (AIMs). The resolution of population ancestry thus depends upon the number of markers, and distinctiveness of the populations involved, which creates tradeoffs for global and local methods of ancestry deconvolution.
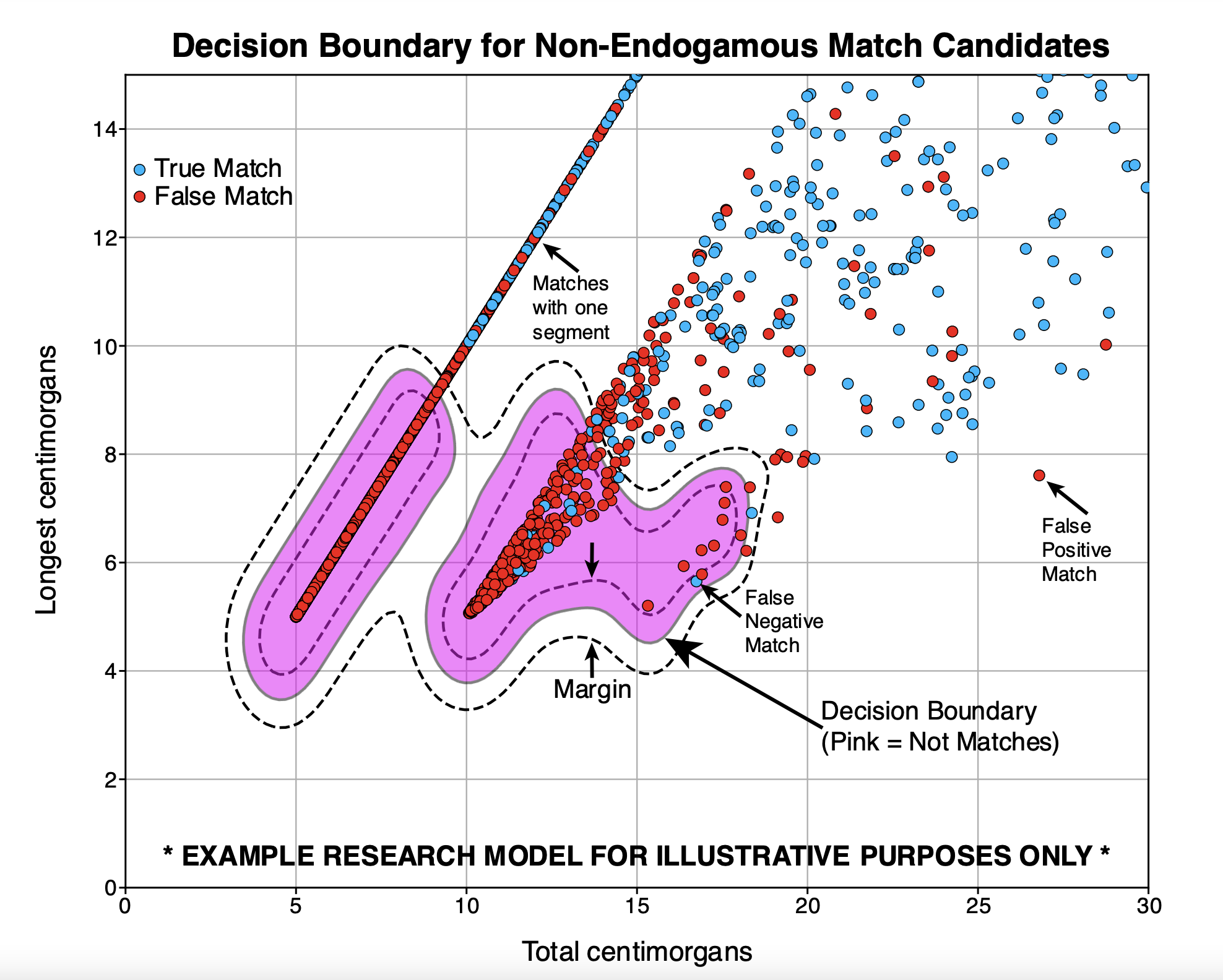
And the other white paper describes the new Family Finder matching algorithm 5.0! This algorithm is a major improvement, by down weighting the influence of so-called “micro-segments.” Here we can see the decision boundary of an SVM model that uses “total cM” and “cM of longest segment” to discern whether a match is real (two individuals sharing a recent common ancestor) or due to isolation by state (IBS). IBS is common due to certain segments being prevalent in the population.
Maier P.A., Hu R., Runfeldt G., Giniebra D., Frichot E. (2021). myOrigins 3.0: Combining global and local methods for determining population ancestry. FamilyTreeDNA White Paper 2021-08-18. [DOI] [PDF]
Hu R., Maier P.A., Runfeldt G., Estes R., Rocha E., Walker A., Baur P., Ty L. (2021). Family Finder Matching 5.0: Matching algorithm and relationship estimation. FamilyTreeDNA White Paper 2021-08-18. [DOI] [PDF]
July 2021
FamilyTreeDNA just released the Chromosome Painter! More will be released soon to explain this exciting new feature, which the Research & Development Team spent a lot of hard work creating.
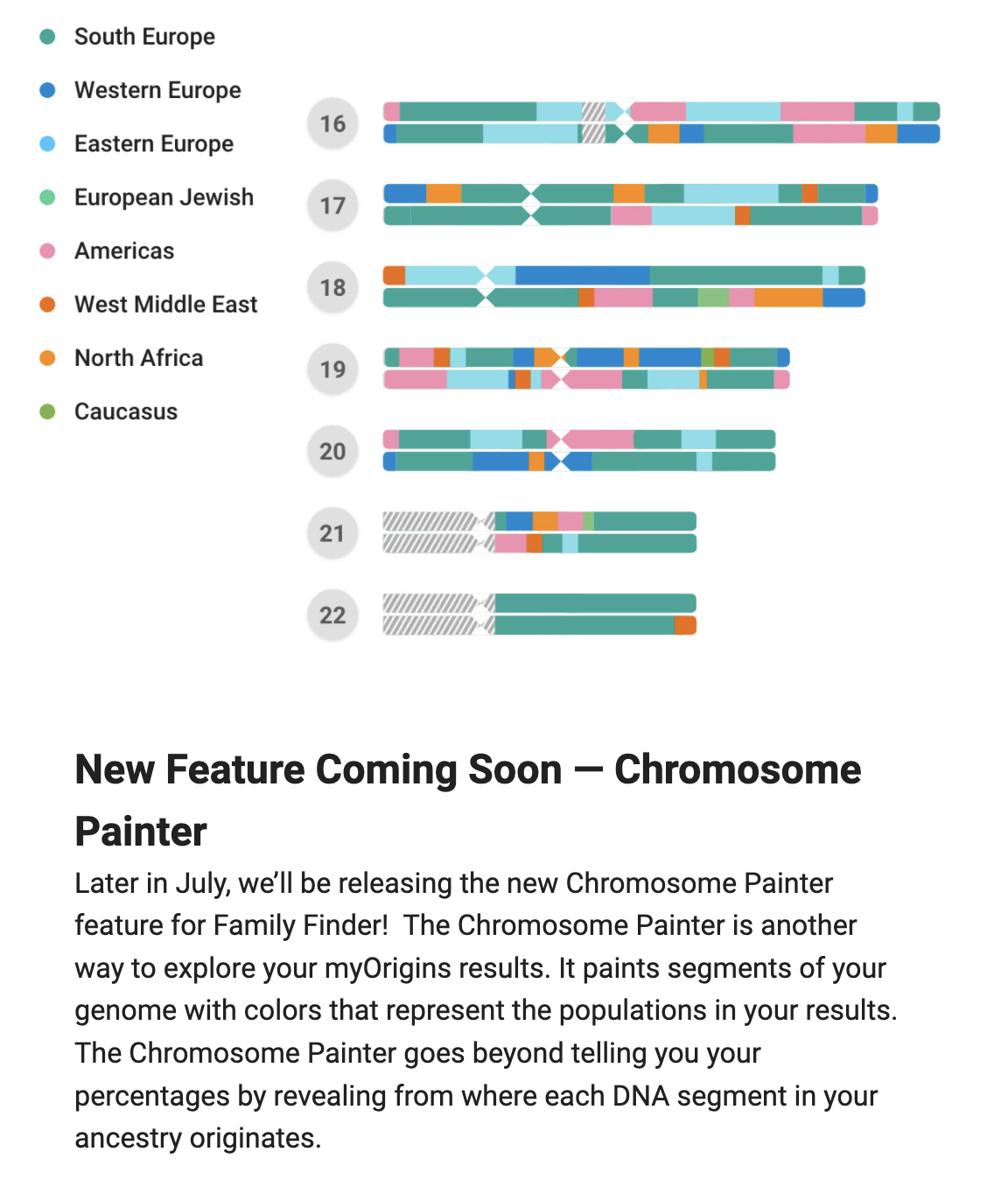
July 2021

My colleague Emily Warshauer and I just published an exciting new article! We used a suite of population genetic analyses to trace the origins of various mutations that cause Recessive Dystrophic Epidermolysis Bullosa (RDEB). We found likely American and European origins, and hint that Sephardic Jews may have carried the mutation to the New World. Another paper we are working on will shed much more light on this story!
Warshauer E., Brown A., Fuentes I., Shortt J., Gignoux C., Montinaro F., Metspalu M., Youssefian L., Vahidnezhad H., Jacków J., Christiano A.M., Uitto J., Fajardo-Ramírez Ó.R., Salas-Alanis J.C., McGrath J.A., Consuegra L., Rivera C., Maier P.A., Runfeldt G., Behar D.M., Skorecki K., Sprecher E., Palisson F., Norris D.A., Bruckner A.L., Kogut I., Bilousova G., Roop D.R. (2021). Ancestral patterns of Recessive Dystrophic Epidermolysis Bullosa mutations in Hispanic populations suggest Sephardic ancestry. American Journal of Medical Genetics Part A, 185(11), 3390–3400. [DOI] [PDF]
April 2021
Miguel Vilar, Roberta Estes, and I gave a really fun and in-depth presentation about human mitochondrial DNA. The hosting institution was Oregon’s chapter of ISOGG. It’s long, but worth it, if you’re interested in either genealogy or mitochondrial evolution. Check it out here:
April 2021
I did a podcast; granted, not my usual MO, but I met the host (Sandy Adams) at a fascinating Egyptology talk by Dr. Salima Ikram, and we decided to chat more (with the camera rolling)! You can check out our conversation here:
September 2020
MyOrigins 3.0 was just released! FamilyTreeDNA increased their count of ancestral reference populations from 24 to a whopping 90! When considering that percentages are estimated for each, this is is basically unparalleled. A huge congratulations to the entire Research & Development Team!!
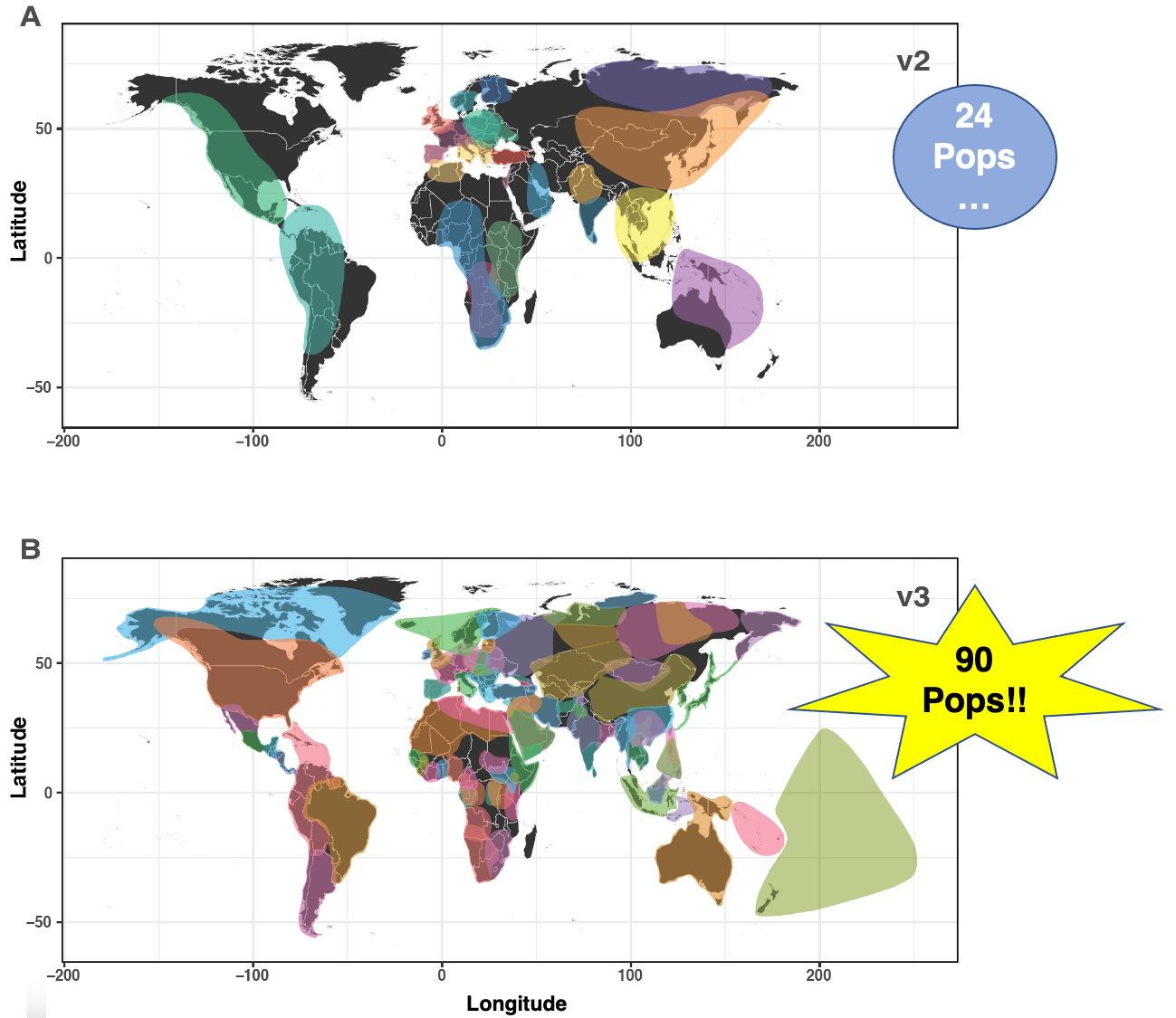
April 2020
My colleague Alexa Lindauer and I just published an exciting new article about the amphibian fungus Bd! We used actual Yosemite toad pond temperature profiles to show decreased fitness of the fungus in a realistic (fluctuating) temperature regime, as compared to lab conditions. We also found that Bd experiences tradeoffs at extreme constant temperatures.
Lindauer A.L., Maier P.A., Voyles J. (2020). Daily fluctuating temperatures decrease growth and reproduction rate of a lethal amphibian fungal pathogen in culture. BMC Ecology, 20(18), 1–9. [DOI] [PDF]
February 2020
RootsTech 2020 was a huge success! We met many enthusiastic genetic genealogists, rolled out several new products and projects, and I gave a presentation about population genetics, and its role in ancestry testing. You can view the presentation here:
November 2019
My collaborator Kai-Hua Jia and I published a new paper using some of the scripting and techniques of my fourth dissertation chapter, to model adaptive genetic response of Chinese arborvitae to climate change!
Jia K.-H., Zhao W., Maier P.A., Hu X.-G., Jin Y., Zhaou S.-S., Jiao S.-Q., El-Kassaby Y.A., Wang T., Wang X.-R., Mao J.-F. (2020). Landscape genomics predicts climate change vulnerability for forest tree: a case from a wide-spread conifer tree, Platycladus orientalis. Evolutionary Applications, 13(4), 665–676. [DOI] [PDF]
October 2019
My first dissertation chapter is now published in Evolution:
Maier P.A., Vandergast A.G., Ostoja S.M., Aguilar A., Bohonak A.J. (2019). Pleistocene glacial cycles drove lineage diversification and fusion in the Yosemite toad (Anaxyrus canorus). Evolution, 73(12), 2476–2496. [DOI] [PDF] [Slides]
In this article, we found that ice ages and physiographic differences between glacial refugia help to shape the phylogenetic structure of an alpine species. Not only that, we discovered several “fused” lineages—toads formed by intraspecific hybridization.
You can get the quick scoop here, with journal club slides posted on FigShare:
January 2019
Another new Natural History Note in Herpetological Review was published! This one describes several observations of tadpoles scavenging on dead or dying animals: trout, their own mother… even each other! Tadpoles are normally detritivores, grazing on algae and potentially ingesting a range of microorganisms. Many species of tadpoles (including Yosemite toads) have been observed supplementing this diet with carrion, but all of the trophic interactions described here were previously unknown.
You can read the article here.

December 2018
A new Natural History Note in Herpetological Review was published! Normally, an excess of males is common amongst pond-breeding amphibians, as strong sexual selection forces males to duke it out for a limited number of available females. This is certainly the usual situation for Yosemite toads. However, this strange observation (from May 2016) shows the opposite: females (one in particular) seem to pester, hit, and jump onto amplectic pairs of toads, seemingly trying to free up a male so she may release her eggs.
The possible implications are discussed: you can read the article here.
September 2018

My dissertation is now published! You can read it on eScholarship or ProQuest (see ‘Publications‘). I will be slowly but surely publishing articles based on the four chapters, which correspond to four evolutionary time frames for the Yosemite toad:
(1) Ancient past: lineage formation and lineage fusion
(2) Recent past: effects of adaptive introgression
(3) Present: population structure and environmentally mediated migration
(4) Future: climate change refugia, and adaptive response to climate change
Maier P.A. (2018). Evolutionary past, present, and future of the Yosemite toad (Anaxyrus canorus): A total evidence approach to delineating conservation units. Ph.D. dissertation. University of California Riverside, and San Diego State University, San Diego, CA. [URL] [DOI] [PDF]
Introduction
Chapter 1: Pleistocene Glacial Cycles Drove Lineage Diversification and Fusion
Chapter 2: Genomic Geography: Islands and Rivers of Divergence at Secondary Contact Zones
Chapter 3: A Novel Genetic Network Model Estimates Environmentally Mediated Migration
Chapter 4: Predicting The Unknown: Conservation Units for Evolutionary Potential
Conclusion